Structural basis of mild developmental delay in the developing human brain connectome
AdaBD made easy
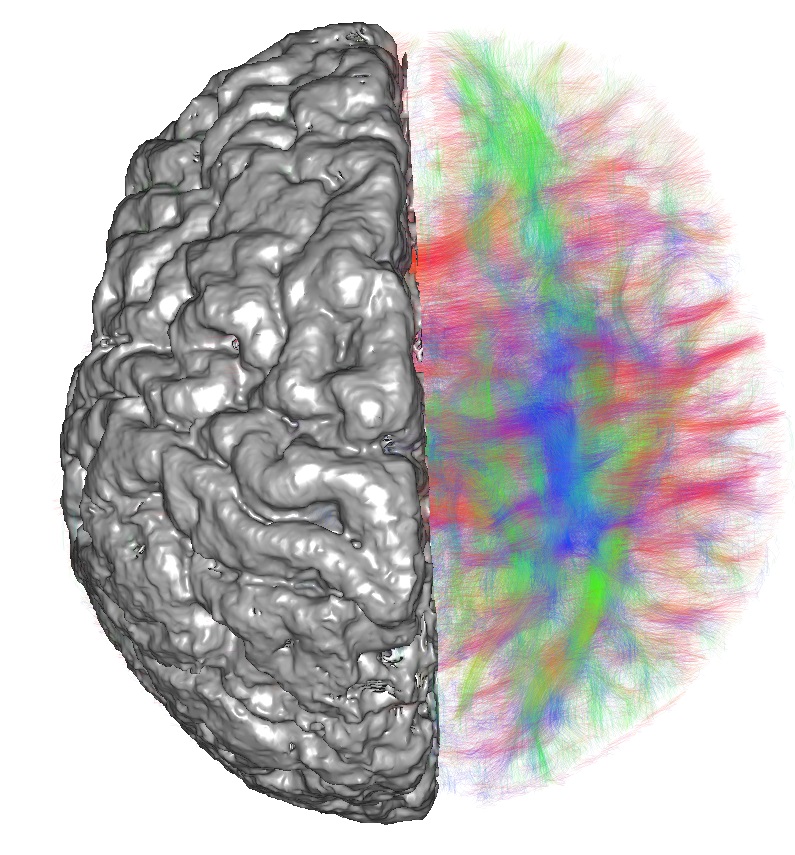
In this project, we developed methods for analysis of existing MRI images from newborns, in order to quantify shape and strengths of brain circuits. We then compared them with the cognitive performance of the children. Our goal is to detect changes in the brain that may be specific for developmental delay disorders.
Research project
Many reasons exist why a child may develop developmental delay (DD) and we do not know yet if they share similar underlying neural conditions, such as deficits in neural circuits. This project aims to characterize the macroscale neuronal circuit structure in children who present mild to moderate DD. We focus on a group of children with mixed reasons for DD, such as congenital heart disorders, prematurity, or congenital central nervous system disorders. We analyzed existing structural and diffusion tensor magnetic resonance imaging (DT-MRI) data from newborns and associated the results with cognitive outcomes. In collaboration with the HDDS platform we implemented analysis techniques that enabled us to look for connectome features that might separate groups (normal vs. pathological). We revealed reduced network integration and increased segregation across the investigated conditions (congenital heart defects, prematurity, spina bifida), although they lacked predictive value for developmental outcome. Furthermore, we developed the neonatal open spina bifida aperta atlas, providing a parcellation strategy for structural connectome analysis and we explored the potential of graph neural networks (GNNs) for neonatal connectome analysis. Our results showed that GNNs were better than traditional models at classifying neonatal brain connectomes based on medical conditions.
The developed analysis pipeline may be applied to additional datasets in the URPP. Moreover, the connectomic framework offers a way for cross-species comparisons, where the anatomical correspondence of regions and networks between species is ambiguous. This will be done in follow-up research projects, such as “Macro-scale connectomic signatures and environmental factors associated with autistic traits in the developing human and mouse brain: a neuroimaging approach” and “Multisensory salience processing in brain circuits of healthy mice and ASD mouse models”.
Project groups
Principal investigator: Andras Jakab, Bea Latal, Michael von Rhein, Valerio Mante
PhD student: Anna Speckert
Platforms: HDDA
Publications
Speckert A, Ji H, Payette K, ..., SPINA BIFIDA STUDY GROUP ZURICH, Jakab A (2024) OSBA: An open neonatal neuroimaging atlas and template for spina bifida aperta. BioRxiv,
Speckert A, Payette K, Knirsch W, von Rhein M, ...,SPINA BIFIDA STUDY GROUP ZURICH, Latal B, Jakab A (2024)Altered connectome topology in newborns at risk for cognitive developmental delay: a cross-etiologic study. bioRxiv,
Ji H, Payette K, Speckert A, ..., Latal B, SPINA BIFIDA STUDY GROUP ZURICH, Jakab A (2024) Thalamic connectivity topography in newborns with spina bifida: association with neurological functional level but not developmental outcome at 2 years Cerebral Cortex bhad438
Payette K, Hongwei L, de Dumast P, Licando E, Ji H, …, Jakab A (2023) Fetal brain tissue annotation and segmentation challenge results Medical Image Analysis 88: 102833
Underlined: AdaBD researchers
